Project informations
- Category: Metabarcoding analysis
- Coordination: Géraldine Pascal
- Project date: on going since 2015
- Project URL: http://frogs.toulouse.inrae.fr
Identify micro-organisms communities in a particular environment using NGS sequences of a targetted gene.
FROGS team:
- Sigenae, GABI, INRAE Jouy en Josas
- NED, GenPhySE, INRAE Toulouse
- Migale, MaIAGE, INRAE Jouy en Josas
- Bioinfo Genotoul, MIAT, INRAE Toulouse
In the past few years there has been a growing interest in the microbiota. Indeed, progress in sequencing has led to important advances in the understanding of the role and the impact of the microbiota on its host. The analysis of microbiomes by the analysis of biomarkers by metabarcoding, such as 16S RNA gene has led us to develop the FROGS software (Escudié et al., 2018).
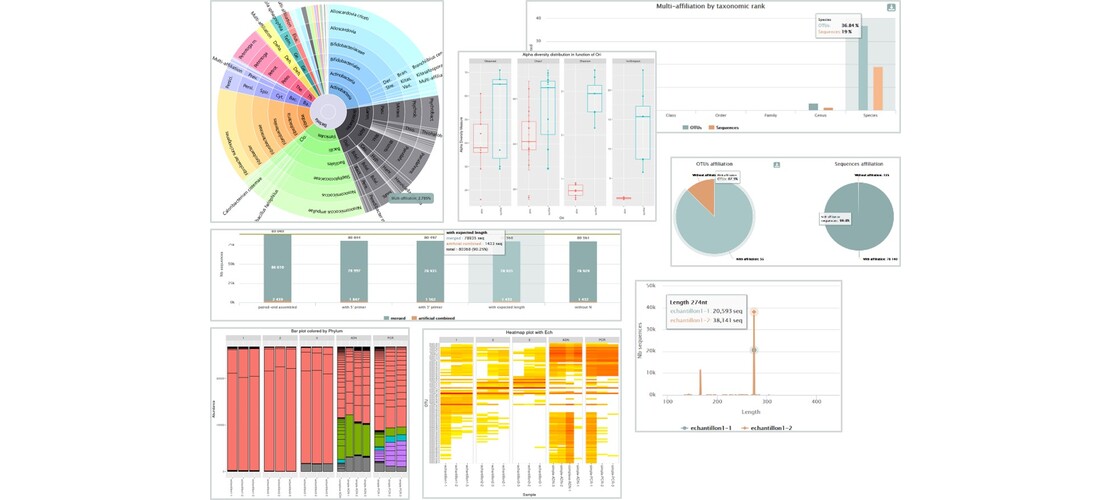
This software allows the analysis of large sets of DNA amplicon sequences with precision and speed. It produces a table of abundance of OTUs (Operational Taxonomic Units) with their taxonomic affiliations. FROGS benefits from the graphical interface of the Galaxy platforms, making it very easy to use. Over the years, FROGS has constantly evolved to allow statistical analysis of data and to meet the challenge of processing ITS sequences (biomarkers of yeasts and fungies). FROGS is the only tool able of managing non-overlapping pairs of reads, as can be the case for ITS markers, RPB2,d1d2, LSU or any other marker larger than 600 nucleotides (which is currently the limit of the paired end short reads). FROGS is widely used by the community and in different context of research, as in :
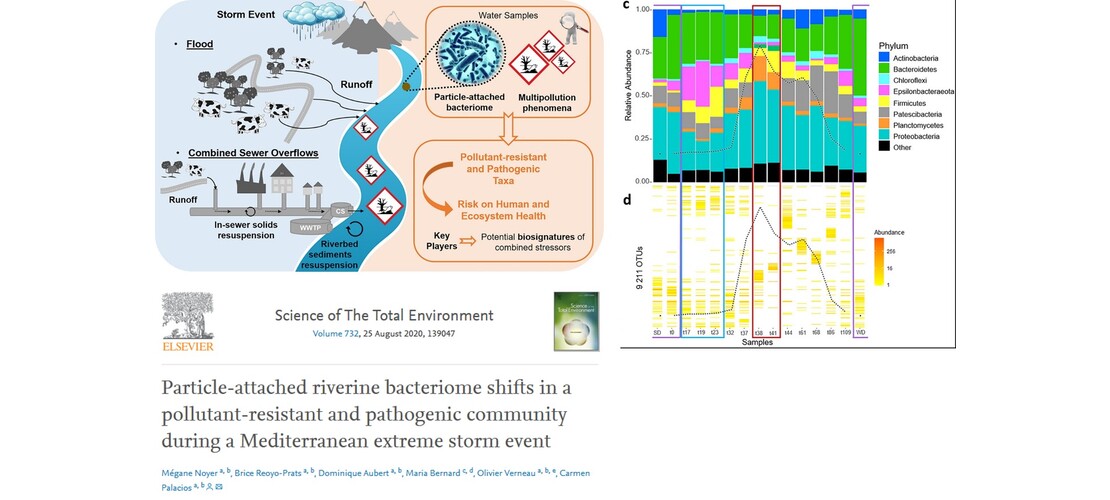
- M. Noyer et al.. Particle-attached riverine bacteriome shifts in a pollutant-resistant and pathogenic community during a Mediterranean extreme storm event, Science of The Total Environment, Volume 732,2020 that studies the microbiome evolution during a flood and its association with pollutant in the river.
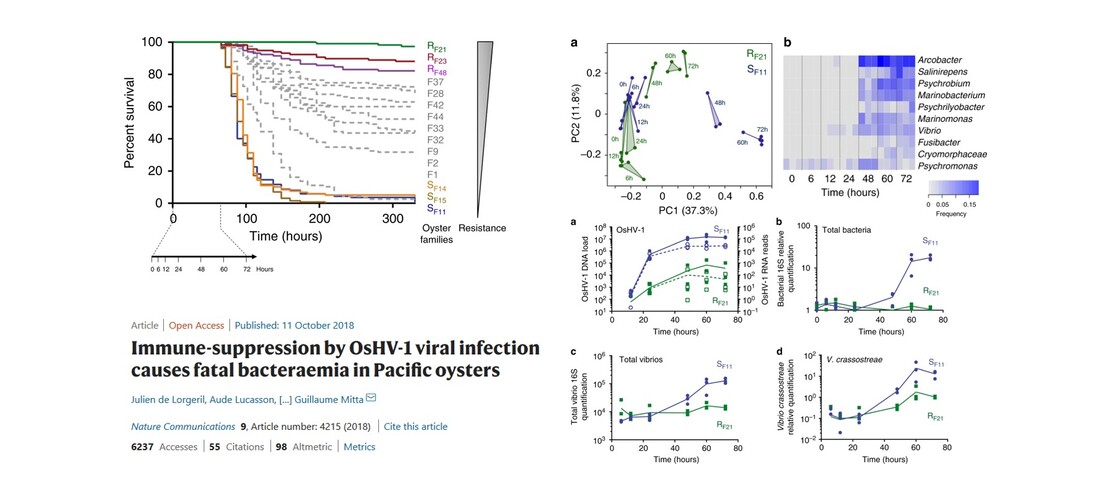
- J. de Lorgeril et al.. Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat Commun 9, 4215 (2018) that studies the microbiome evolution during oyster infection by the Ostreid herpesvirus OsHV-1 µVar.
FROGS in Brief:
- Analyse a huge amount of sequence with precision and in reasonable time.
- Generate OTU abundance table with taxonomic affiliations
- Usable in command line or under Galaxy plateform
- Composed of 25 bioinformatics and statistical tools
- Analyse long amplicon sequences as ITS (for yeast or fungi) by dealing with non-overlapping read pairs (ITS, RPB2, d1d2, LSU ...)
- Acclaimed by the community:
- more than 4200 downloads on anaconda.org/bioconda/frogs
- more than 200 citations of the publication
- At least, 13 Galaxy server proposed FROGS
- more than 24,000 visitors on the FROGS website
The FROGS team provide around 5 trainings per year at Toulouse and Jouy-en-Josas INRAE centres. Please, refer to the Bioinfo Genotoul trainings and Migale trainings web pages for more information.