About Us
Sigenae - an INRAE bioinformatic platform
Sigenae is an INRAE bioinformatic platform specialized in animal genomic data processing. Seven engineers located in Toulouse and Jouy-en-Josas help biologists to extract scientific knowledge from datasets generated by high-throughput sequencers and genotypers.
- The team takes part in different national and international projects such as
1000 bull genomes,
1000 goat genomes,
FAANG,
BovReg,
STURGEoNOMICS,
GenoFish.
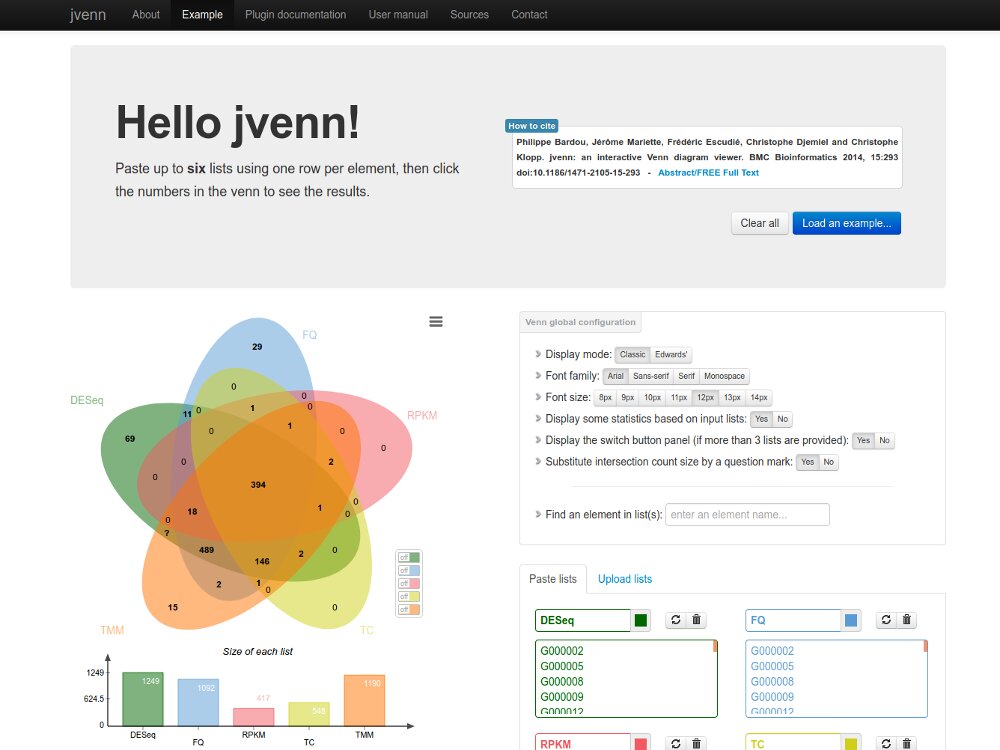
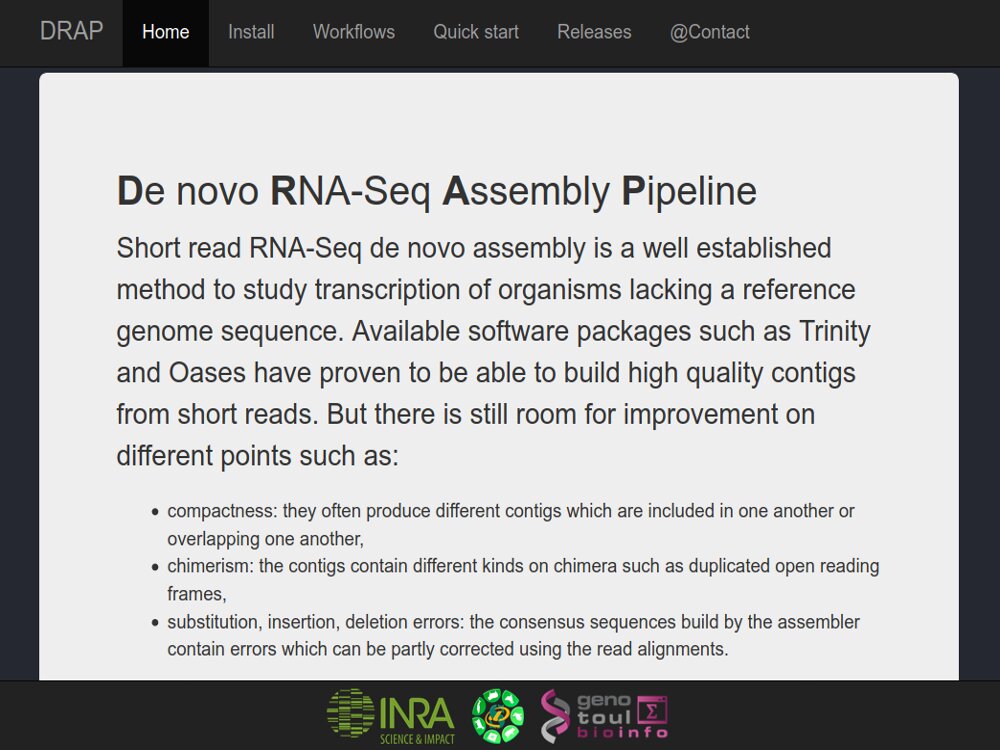
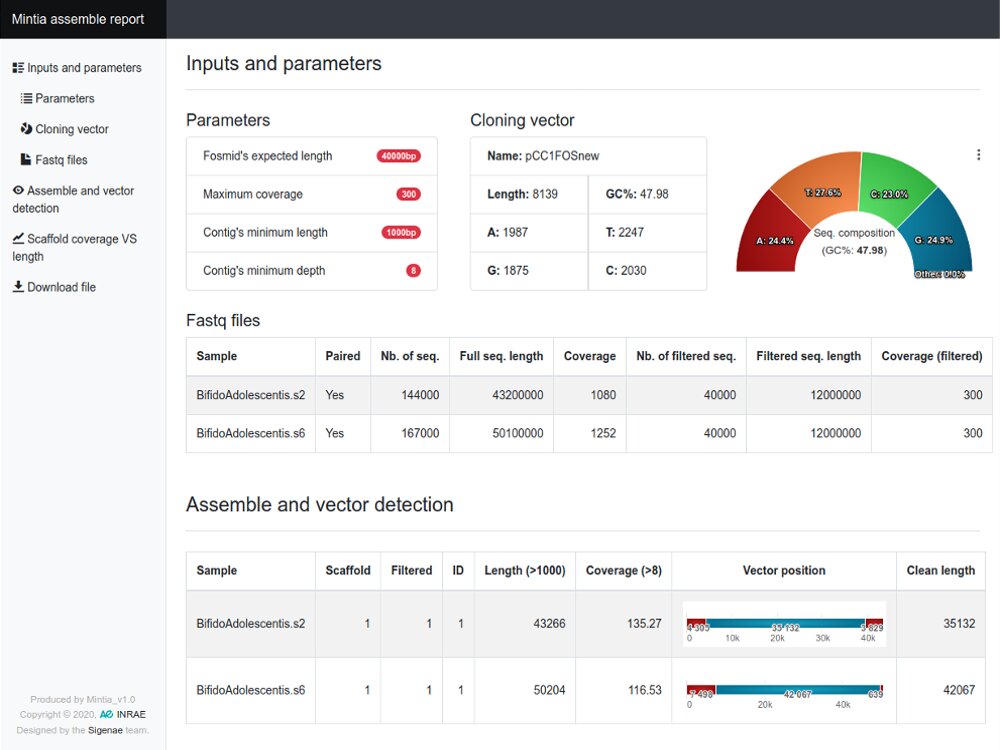
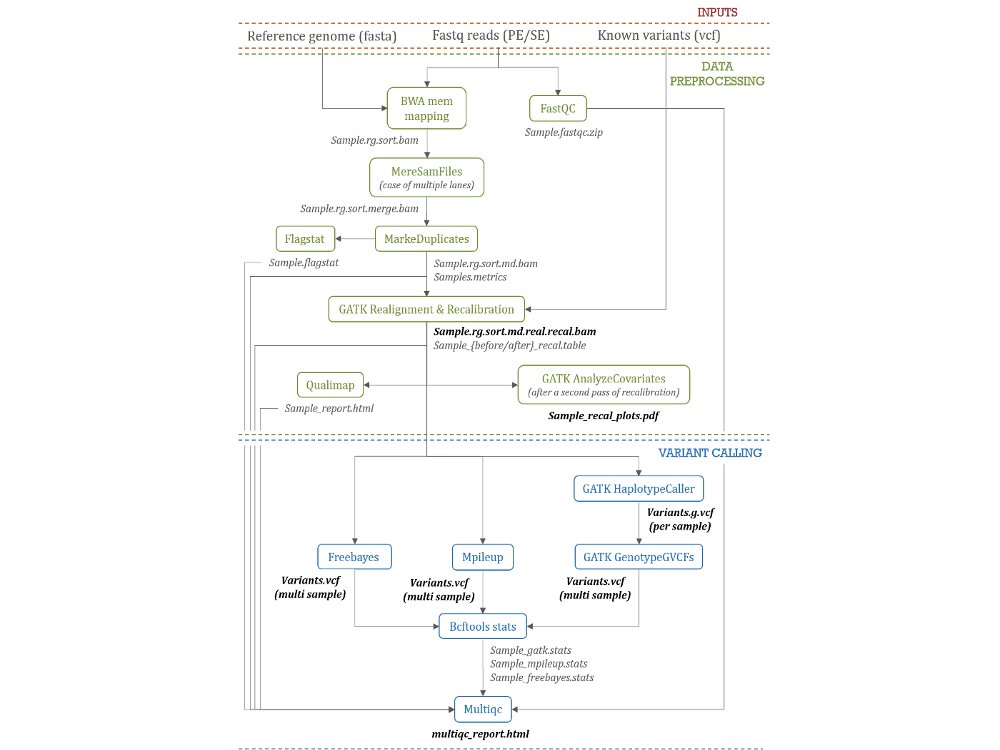
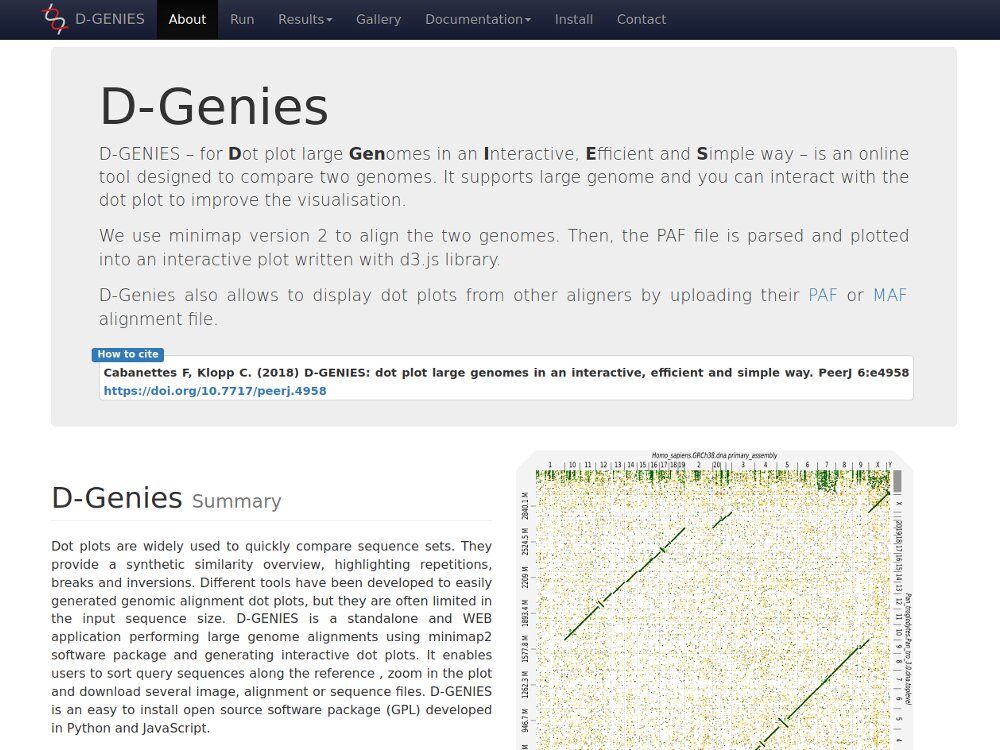
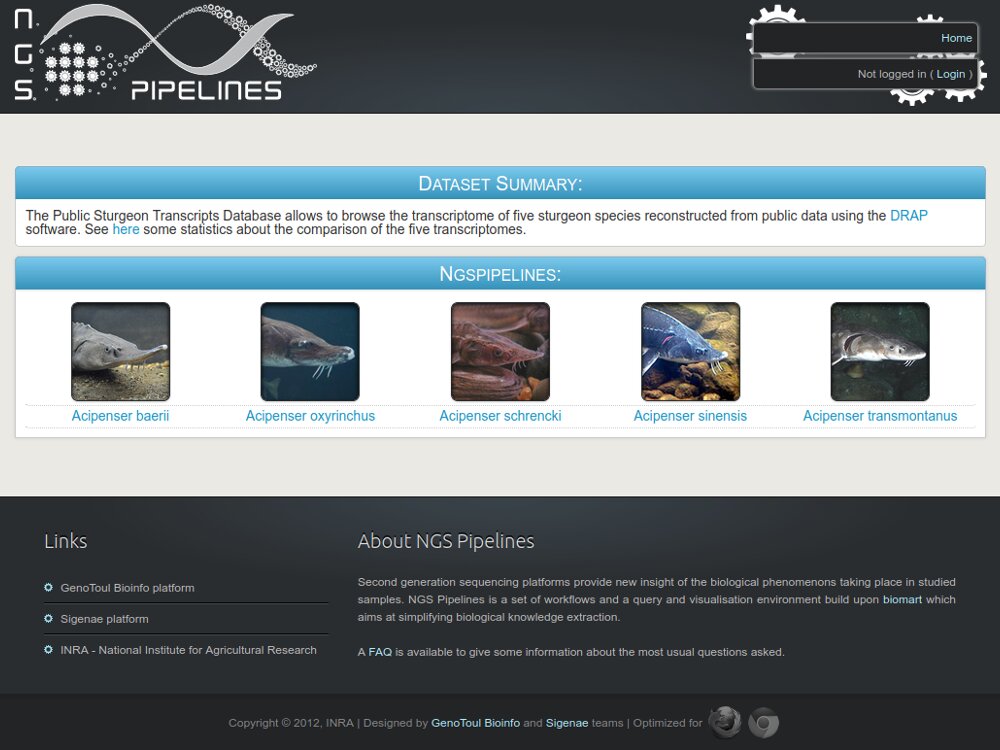
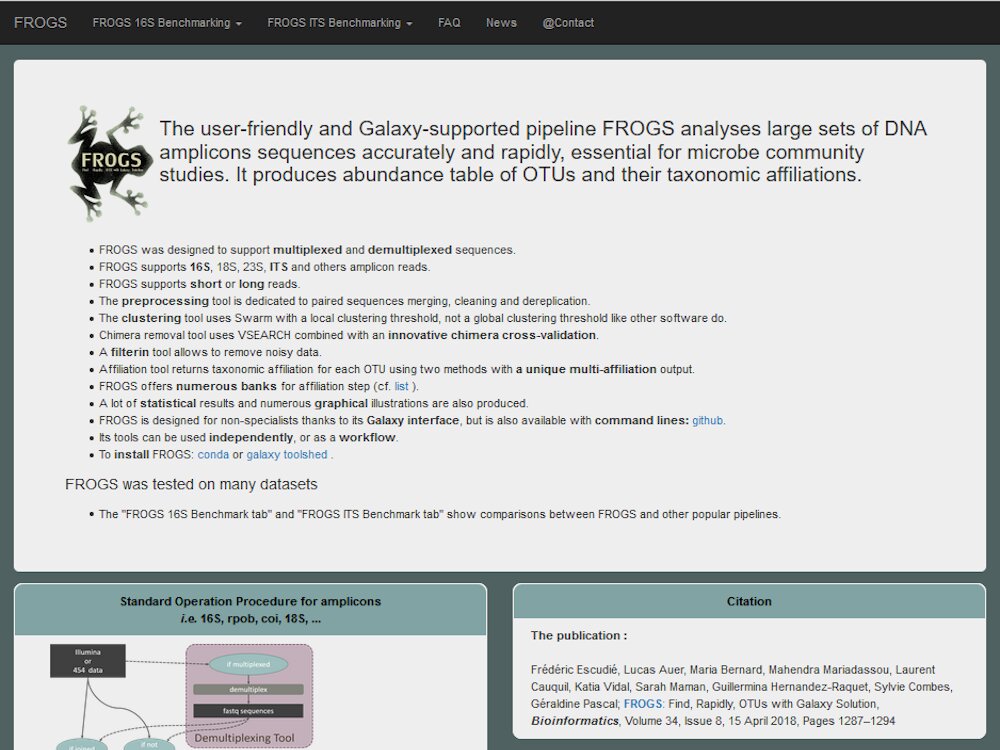
Our main contributions are data processing, training and software development. - We have participated in and published open source software packages such as FROGS, jVenn, DRAP, D-Genies, NG6, RNAbrowse.
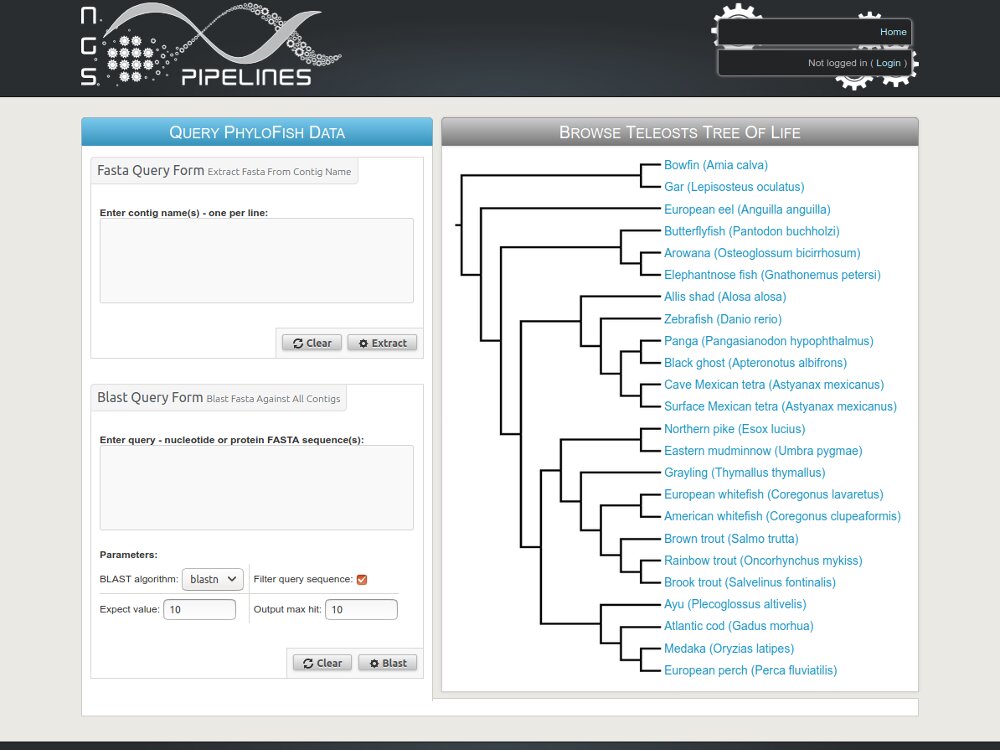
- We also maintain published databases like Phylofish and SSturgeon.
The team members co-authored numerous scientific publications.
Project support
The Sigenae team are involved in many projects.
See below, by topic, some examples of most significant supports.
Variant detection and annotation
Example with the 1000 goat genomes project - VarGoats...
De novo assembly and annotation of genomes
Example with the GenoFish project...
16S diversity
Example with metabarcoding analysis project - FROGS...
RNA-Seq transcriptomic analyses
Examples with recent projects...
System and software engineering
Example with Galaxy...
Trainings
The members of Sigenae are involved in numerous training courses...
Development project
The Sigenae team develops or is involved in the development of many tools,
databases and workflows.
See below some examples of significant achievements.
- All
- Tools
- Databases
- Workflows

Christophe Klopp
Project coordinatorGenomic, Transcriptomic, Assembly
Data processing, AWK, Perl, Python, R.

Philippe Bardou
Bioinformatics engineerDNAseq, Variant calling, sRNAseq (miRNA)
Perl, Python, GIT, HTML5, CSS3, Javascript.

Maria Bernard
Bioinformatics engineerVariant calling, RADseq, Metagenomic, RNASeq, ASE
Python, Snakemake, GIT, Rmarkdown, Galaxy.

Cedric Cabau
Bioinformatics engineerRNAseq, Assembly, Annotation
Perl, Awk, Python, GIT, HTML/CSS/JS.

Mathieu Charles
Bioinformatics engineerVariant calling, CNV, RNAseq
Perl, Python, Java, C++, Snakemake, Jupyter, R.

Patrice Dehais
Sysadmin and B.E.Sysadmin HPC Virtualization Parallelization/Optimization applied to bioinformatics software.

Cervin Guyomar
Bioinformatics engineerGenomics, metagenomics, functional genomics
R, Python, NextFlow.

Sarah Maman
Bioinformatics engineerRNAseq, Galaxy, elearning
Perl, Python, Cheetah, Nextflow.

Odile Rogier
Bioinformatics engineerVariant calling, RNAseq
GIT, Python
Frequently Asked Questions
-
What is Sigenae?
Sigenae is a bioinformaticians team providing services to INRAE aquaculture and farm animal biologists.
The Sigenae platform is part of the BioinfOmics Research Infrastructure of INRAE for bioinformatics. -
Who do we collaborate with?
Research teams of french research institutes: INRAE, CNRS, ENS, Ifremer, CIRAD, ENVT, Université de Perpignan. Among foreign parterns, we collaborate with researchers from the University of Würzburg (Germany), University of Montevideo (Uruguay), University of Szczecin (Poland), University of Lauzanne (Switzerland), Institute of Animal Reproduction and Food in Olsztyn (Poland), USDA Agricultural Research Service in Stoneville (USA), Leibniz-Institute of Freshwater Ecology and Inland Fisheries (Germany).
-
How to interact with us?
For any request please use this link to contact the Sigenae team.
If you wish to contact any of us directly, please see the contact section below. -
How to cite us?
Depending on the help provided you can cite us in acknowledgements, references or both.
Example for acknowledgements: we wish to thank the Sigenae group for ....
Affiliations for references:
C. Klopp, Q. Boone Sigenae, MIAT UR875, INRAE, F-31326, Castanet Tolosan, France. M. Bernard, M. Charles Sigenae, GABI, INRAE, AgroParisTech, Université Paris-Saclay, Jouy-en-Josas, France. P. Bardou, C. Cabau, P. Dehais, Sigenae, GenPhySE, Université de Toulouse, INRAE, ENVT, F-31326, Castanet Tolosan, France. C. Guyomar, S. Maman - How can I get the Sigenae logo?
Contact
The members of the group are located in two sites (Jouy-en-josas and Toulouse).
For any request please use this
link
to contact us.
Location:
Centre de recherche Occitanie-Toulouse
24, Chemin de Borde Rouge - Auzeville
CS 52627
31326 Castanet-Tolosan Cedex, FRANCE
Members:
| First Name | Last Name | Phone Number | |
|---|---|---|---|
| Christophe | Klopp | +335 61 28 50 36 | |
| Philippe | Bardou | +335 61 28 57 09 | |
| Cédric | Cabau | +335 61 28 54 60 | |
| Patrice | Dehais | +335 61 28 57 08 | |
| Cervin | Guyomar | +335 61 28 51 17 | |
| Sarah | Maman | +335 61 28 57 08 |
Location:
Centre INRAE de Jouy en Josas
Domaine de Vilvert
4 Avenue Jean Jaurès
F-78352 JOUY EN JOSAS Cedex, FRANCE
Members:
| First Name | Last Name | Phone Number | |
|---|---|---|---|
| Maria | Bernard | +331 34 65 25 77 | |
| Mathieu | Charles | +331 34 65 28 15 | |
| Odile | Rogier |